Panpipes - multimodal single cell pipelines
What is Panpipes?
Panpipes is a collection of cgat-core/ruffus pipelines to streamline the analysis of multi-modal single cell data. Panpipes supports any combination of the following single-cell modalities: scRNAseq, CITEseq, scV(D)Jseq, and scATACseq

Check out the installation and usage guidelines page for further information.
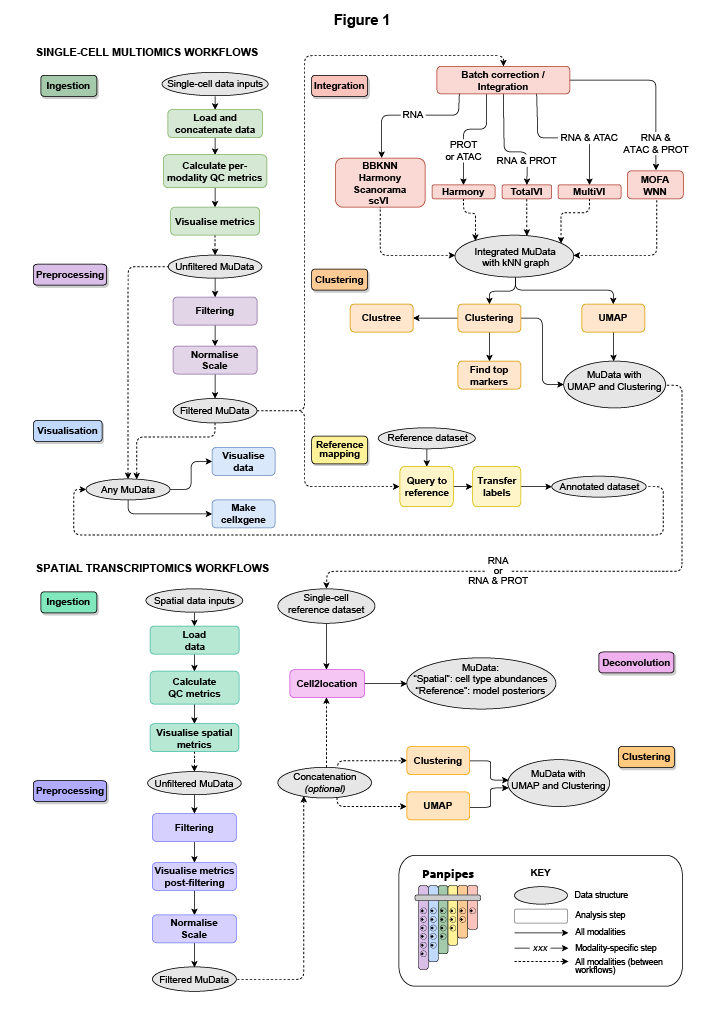
Available workflows for multimodal data:
Ingestion and Quality Control metrics generation : for the ingestion of data and computation of QC metrics
Preprocessing : for filtering and normalizing each modality
Integration: integrate and batch correction using single and multimodal methods
Clustering : cell clustering on single modalities
Reference Mapping (refmap) : transfer scvi-tools models from published data to your data
Visualization : visualize metrics from other pipelines in the context of experiment metadata
Available workflows for spatial data:
Ingesting spatial data : for the ingestion of spatial transcriptomics (ST) data (Vizgen, Visium) and computation of QC metrics
Preprocessing spatial data: for filtering and normalizing ST data
Deconvoluting spatial data : for the cell type deconvolution of ST slides
Clustering spatial data : for clustering ST data